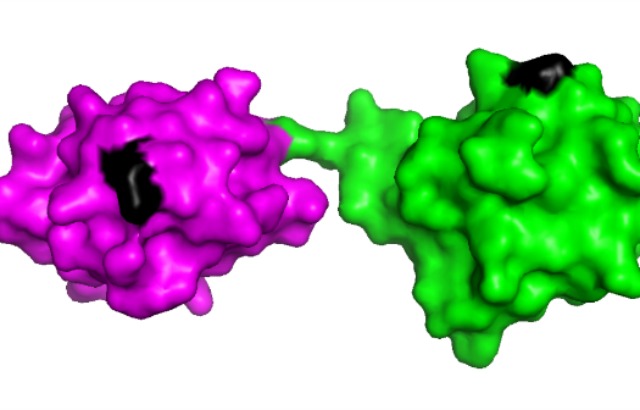
Schematic model of two linearly linked ubiquitin molecules. Credit: Koraljka Husnjak using PyMOL software
Ubiquitin is a small protein that controls and modulates the function of other cellular proteins by connecting to them. The attachment occurs in many different forms, either as single molecule or in the form of distinct ubiquitin chains, leading to diverse conformations and varying cellular outcomes. Scientists often refer to this as the secret ubiquitin code, which still needs to be fully deciphered.
Recently, scientists discovered that ubiquitin molecules are not only assembled in a non-linear manner, but also build linear chains, in which the head of one ubiquitin is linked to the tail of another ubiquitin molecule. So far, only two highly specific enzymes are known to be capable of synthesizing and degrading such linear ubiquitin chains. However, target proteins of linear ubiquitination, as well as their specific cellular functions, have largely remained elusive.
The novel technology, published in the journal Nature Methods, enables the systematic analysis of linear ubiquitination targets.
“The slow progress in this research area was mainly due to the lack of suitable methods for proteomic analysis of proteins modified with linear ubiquitin chains”, explained lead author Koraljka Husnjak from Goethe University Frankfurt. Her team solved the problem by internally modifying the ubiquitin molecule in such a way that it maintains its cellular functions whilst at the same time enabling the enrichment and further analysis of linear ubiquitin targets by mass spectrometry.
Modified form of the ubiquitin molecule
Co-author Dr Benjamin Stieglitz from QMUL’s School of Biological and Chemical Sciences (SBCS) says: “This work was a great collaborative effort in which research labs with complementary expertise teamed up to find a new way to detect proteins which are modified with linear polyubiquitin. The centrepiece of the new method is a modified form of the ubiquitin molecule.”
“Our part was to show that the modification does not affect the three-dimensional shape of ubiquitin. The analysis of the molecular structure confirmed that this is indeed not the case, which indicates that the new method can be applied without any limitations,” he added.
The technology makes it possible to identify target proteins modified by linear ubiquitin, and to detect the exact position within the protein where the linear chain is attached.
Errors in the ubiquitin system have been linked to numerous diseases including cancer and neurodegenerative disorders such as Parkinson’s disease, but also to the development and progression of infections and inflammatory diseases.



