Enzyme Nomenclature. Recommendations 1992
Continued from EC 3.4.21.1 to EC 3.4.21.50
EC 3.4.21 (Continued)
Serine endopeptidases
Contents
EC 3.4.21.51 Deleted entry
EC 3.4.21.52 Deleted entry:
EC 3.4.21.53 endopeptidase La
EC 3.4.21.54 γ-renin
EC 3.4.21.55 venombin AB
EC 3.4.21.56 Deleted entry:
EC 3.4.21.57 leucyl endopeptidase
EC 3.4.21.58 Deleted entry:
EC 3.4.21.59 tryptase
EC 3.4.21.60 scutelarin
EC 3.4.21.61 kexin
EC 3.4.21.62 subtilisin
EC 3.4.21.63 oryzin
EC 3.4.21.64 peptidase K
EC 3.4.21.65 thermomycolin
EC 3.4.21.66 thermitase
EC 3.4.21.67 endopeptidase So
EC 3.4.21.68 t-plasminogen activator
EC 3.4.21.69 activated protein C (thrombin-activated peptidase)
EC 3.4.21.70 pancreatic endopeptidase E
EC 3.4.21.71 pancreatic elastase II
EC 3.4.21.72 IgA-specific serine endopeptidase
EC 3.4.21.73 u-plasminogen activator
EC 3.4.21.74 venombin A
EC 3.4.21.75 furin
EC 3.4.21.76 myeloblastin
EC 3.4.21.77 semenogelase
EC 3.4.21.78 granzyme A
EC 3.4.21.79 granzyme B
EC 3.4.21.80 streptogrisin A
EC 3.4.21.81 streptogrisin B
EC 3.4.21.82 glutamyl endopeptidase II
EC 3.4.21.83 oligopeptidase B
EC 3.4.21.84 limulus clotting factor 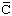
EC 3.4.21.85 limulus clotting factor 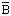
EC 3.4.21.86 limulus clotting enzyme
EC 3.4.21.87 omptin
EC 3.4.21.87 now EC 3.4.23.49
EC 3.4.21.88 repressor LexA
EC 3.4.21.89 signal peptidase I
EC 3.4.21.90 togavirin
EC 3.4.21.91 flavivirin
EC 3.4.21.92 endopeptidase Clp
EC 3.4.21.93 proprotein convertase 1
EC 3.4.21.94 proprotein convertase 2
EC 3.4.21.95 snake venom factor V activator
EC 3.4.21.96 lactocepin
EC 3.4.21.97 assemblin
EC 3.4.21.98 hepacivirin
EC 3.4.21.99 spermosin
EC 3.4.21.100 sedolisin
See the following file for:
EC 3.4.21.101 to EC 3.4.21.123
Entries
[EC 3.4.21.51 Deleted entry: Leukocyte-membrane neutral endopeptidase (EC 3.4.21.51 created 1984, deleted 1992)]
[EC 3.4.21.52 Deleted entry: Cathepsin R (EC 3.4.21.52 created 1981 as EC 3.4.99.33, transferred 1984 to EC 3.4.21.52, deleted 1992)]
EC 3.4.21.53
Accepted name: endopeptidase La
Reaction: Hydrolysis of proteins in presence of ATP
Other names: ATP-dependent serine proteinase; lon proteinase; protease La; proteinase La; ATP-dependent lon proteinase; ATP-dependent protease La; Escherichia coli proteinase La; Escherichia coli serine proteinase La; gene lon protease; gene lon proteins; PIM1 protease; PIM1 proteinase; serine protease La
Comments: Product of the lon gene in Escherichia coli. ATP hydrolysis is linked with peptide bond hydrolysis; vanadate inhibits both reactions. Type example of peptidase family S16. A similar enzyme occurs in animal mitochondria
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 79818-35-2
References
1. Desautels, M. and Goldberg, A.L. Demonstration of an ATP-dependent, vanadate-sensitive endoprotease in the matrix of rat liver mitochondria. J. Biol. Chem. 257 (1982) 11673-11679. [PMID: 6749845]
2. Larimore, F.S., Waxman, L. and Goldberg, A.L. Studies of the ATP-dependent proteolytic enzyme, protease La, from Escherichia coli. J. Biol. Chem. 257 (1982) 4187-4195. [PMID: 7040380]
3. Chin, D.T., Goff, S.A., Webster, T., Smith, T. and Goldberg, A.L. Sequence of the lon gene in Escherichia coli. A heat-shock gene which encodes the ATP-dependent protease La. J. Biol. Chem. 263 (1988) 11718-11728. [PMID: 3042779]
[EC 3.4.21.53 created 1986]
EC 3.4.21.54
Accepted name: γ-renin
Reaction: Cleavage of the Leu Leu bond in synthetic tetradecapeptide renin substrate (horse), to produce angiotensin I, but not active on natural angiotensinogen, unlike renin (EC 3.4.23.15). Also hydrolyses Bz-Arg-p-nitroanilide
Leu bond in synthetic tetradecapeptide renin substrate (horse), to produce angiotensin I, but not active on natural angiotensinogen, unlike renin (EC 3.4.23.15). Also hydrolyses Bz-Arg-p-nitroanilide
Comments: A member of the tissue kallikrein family, from submandibular glands of male mice. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number: 85270-20-8
References
1. Poe, M., Wu, J.K., Florance, J.R., Rodkey, J.A., Bennett, C.D. and Hoogsteen, K. Purification and properties of renin and γ-renin from the mouse submaxillary gland. J. Biol. Chem. 258 (1983) 2209-2216[PMID: 6337154]
2. Drinkwater, C.C., Evans, B.A. and Richards, R.I. Sequence and expression of mouse γ-renin. J. Biol. Chem. 263 (1988) 8565-8568. [PMID: 3288617]
[EC 3.4.21.54 created 1986]
EC 3.4.21.55
Accepted name: venombin AB
Reaction: Selective cleavage at Arg bonds in fibrinogen to form fibrin and release fibrinopeptides A and B
bonds in fibrinogen to form fibrin and release fibrinopeptides A and B
Other names: gabonase; okinaxobin II; Bitis gabonica venom serine proteinase; afaâcytin
Comments: From the venom of the Gaboon viper Bitis gabonica. Activates Factor XIII. Not inhibited by antithrombin III/heparin or hirudin, unlike EC 3.4.21.5, thrombin
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number: 104003-74-9
References
1. Pirkle, H., Theodor, I., Miyada, D. and Simmons, G. Thrombin-like enzyme from the venom of Bitis gabonica. Purification, properties and coagulant actions. J. Biol. Chem. 261 (1986) 8830-8835. [PMID: 3522580]
[EC 3.4.21.55 created 1989]
[EC 3.4.21.56 Deleted entry: Euphorbain. Now considered EC 3.4.21.25, cucumisin (EC 3.4.21.56 created 1972 as EC 3.4.99.7, transferred 1989 to EC 3.4.21.56, deleted 1992)]
EC 3.4.21.57
Accepted name: leucyl endopeptidase
Reaction: Hydrolysis of proteins. Preferential cleavage: Leu in small molecule substrates
in small molecule substrates
Other names: plant Leu-proteinase; leucine-specific serine proteinase; leucine endopeptidase; spinach serine proteinase (leucine specific); spinach leucine-specific serine proteinase; Leu-proteinase
Comments: From leaves of the spinach plant (Spinacia oleracea)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number: 136396-22-0
References
1. Aducci, P., Ascenzi, P., Pierini, M. and Ballio, A. Purification and characterization of Leu-proteinase, the leucine specific serine proteinase from spinach (Spinacia oleracea L.) leaves. Plant Physiol. 81 (1986) 812-816
2. Aducci, P., Ascenzi, P. and Ballio, A. Esterolytic properties of leucine-proteinase, the leucine-specific serine proteinase from spinach (Spinacia oleracea L.). Plant Physiol. 82 (1986) 591-593
[EC 3.4.21.57 created 1989]
[EC 3.4.21.58 Deleted entry: prohormone serine proteinase (EC 3.4.21.58 created 1989, deleted 1992)]
EC 3.4.21.59
Accepted name: tryptase
Reaction: Preferential cleavage: Arg , Lys
, Lys , but with more restricted specificity than trypsin
, but with more restricted specificity than trypsin
Other names: mast cell tryptase; mast cell protease II; skin tryptase; lung tryptase; pituitary tryptase; mast cell neutral proteinase; mast cell tryptase; mast cell neutral proteinase; mast cell serine proteinase II; mast cell proteinase II; mast cell serine proteinase tryptase; rat mast cell protease II; tryptase M
Comments: Occurs as a tetrameric molecule with high affinity for heparin, in mast cell granules. In peptidase family S1 (trypsin family). Not inhibited by α1-proteinase inhibitor or α2-macroglobulin
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 97501-93-4
References
1. Tanaka, T., McRae, B.J., Cho, K., Cook, R., Fraki, J.E., Johnson, D.A. and Powers, J.C. Mammalian tissue trypsin-like enzymes. Comparative reactivities of human skin tryptase, human lung tryptase, and bovine trypsin with peptide 4-nitroanilide and thioester substrates. J. Biol. Chem. 258 (1983) 13552-13557. [PMID: 6358206]
2. Kido, H., Fukusen, N. and Katunuma, N. Chymotrypsin- and trypsin-type serine proteases in rat mast cells: properties and functions. Arch. Biochem. Biophys 239 (1985) 436-443. [PMID: 3890754]
3. Cromlish, J.A., Seidah, N.G., Marcinkiewitz, M., Hamelin, J., Johnson, D.A. and Chrétien, M. Human pituitary tryptase: molecular forms, NH2-terminal sequence, immunocytochemical localization, and specificity with prohormone and fluorogenic substrates. J. Biol. Chem. 262 (1987) 1363-1373. [PMID: 3543004]
4. Harvima, I.T., Schechter, N.M., Harvima, R.J. and Fräki, J.E. Human skin tryptase: purification, partial characterization and comparison with human lung tryptase. Biochim. Biophys. Acta 957 (1988) 71-80. [PMID: 3140898]
5. Vanderslice, P., Ballinger, S.M., Tam, E.K., Goldstein, S.M., Craik, C.S. and Caughey, G. Human mast cell tryptase: multiple cDNAs and genes reveal a multigene serine protease family. Proc. Natl. Acad. Sci. USA 87 (1990) 3811-3815. [PMID: 2187193]
[EC 3.4.21.59 created 1992]
EC 3.4.21.60
Accepted name: scutelarin
Reaction: Selective cleavage of Arg Thr and Arg
Thr and Arg Ile in prothrombin to form thrombin and two inactive fragments
Ile in prothrombin to form thrombin and two inactive fragments
Other name(s): taipan activator; Oxyuranus scutellatus prothrombin-activating proteinase
Comments: From the venom of the Taipan snake (Oxyuranus scutellatus). Converts prothrombin to thrombin. Specificity is similar to that of Factor Xa (EC 3.4.21.6). However, unlike Factor Xa this enzyme can cleave its target in the absence of coagulation Factor Va. Activity is potentiated by phospholipid and Ca2+ which binds via γ-carboxyglutamic acid residues. Similar enzymes are known from the venom of other Australian elapid snakes, including Pseudonaja textilis textilis. Oxyuranus microlepidotus and Demansia nuchalis affinis.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number: 93389-45-8
References:
1. Walker, F.J., Owen, W.G. and Esmon, C.T. Characterization of the prothrombin activator from the venom of Oxyuranus scutellatus scutellatus (taipan venom). Biochemistry 19 (1980) 1020-1023. [PMID: 6986908]
2. Speijer, H., Govers-Reimslag, J.W., Zwaal, R.F. and Rosing, J. Prothrombin activation by an activator from the venom of Oxyuranus scutellatus (taipan snake). J. Biol. Chem. 261 (1986) 13258-13267. [PMID: 3531198]
[EC 3.4.21.60 created 1978 as EC 3.4.99.28, transferred 1992 to EC 3.4.21.60, modified 2011]
EC 3.4.21.61
Accepted name: kexin
Reaction: Cleavage of -Lys-Arg and -Arg-Arg
and -Arg-Arg bonds to process yeast α-factor pheromone and killer toxin precursors
bonds to process yeast α-factor pheromone and killer toxin precursors
Other names: yeast KEX2 protease; proteinase yscF; prohormone-processing endoprotease; paired-basic endopeptidase; yeast cysteine proteinase F (misleading); paired-basic endopeptidase; andrenorphin-Gly-generating enzyme; endoproteinase Kex2p; gene KEX2 dibasic proteinase; Kex 2p proteinase; Kex2 endopeptidase; Kex2 endoprotease; Kex2 endoproteinase; Kex2 protease; proteinase Kex2p; Kex2-like precursor protein processing endoprotease; prohormone-processing KEX2 proteinase; prohormone-processing proteinase; proprotein convertase; protease KEX2; Kex2 proteinase; Kex2-like endoproteinase
Comments: A Ca2+-activated peptidase of peptidase family S8, containing Cys near the active site His, and inhibited by p-mercuribenzoate. Similar enzymes occur in mammals. Formerly EC 3.4.22.23
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 99676-46-7
References
1. Julius, D., Brake, A., Blair, L., Kunisawa, R. and Thorner, J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-α-factor. Cell 37 (1984) 1075-1089. [PMID: 6430565]
2. Achstetter, T. and Wolf, D.H. Hormone processing and membrane-bound proteinases in yeast. EMBO J. 4 (1985) 173-177. [PMID: 3894003]
3. Mizuno, K., Nakamura, T., Ohshima, T., Tanaka, S. and Matsuo, H. Yeast KEX2 gene encodes an endopeptidase homologous to subtilisin-like serine proteases. Biochem. Biophys. Res. Commun. 156 (1988) 246-254 [PMID: 2845974]
4. Fuller, R.S., Brake, A. and Thorner, J. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc. Natl. Acad. Sci. USA 86 (1989) 1434-1438. [PMID: 2646633]
5. Mizuno, K., Nakamura, T., Ohshima, T., Tanaka, S. and Matsuo, H. Characterization of KEX2-encoded endopeptidase from yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 159 (1989) 305-311 [PMID: 2845974]
[EC 3.4.21.61 created 1989 as EC 3.4.22.23, transferred 1992 to EC 3.4.21.61]
EC 3.4.21.62
Accepted name: subtilisin
Reaction: Hydrolysis of proteins with broad specificity for peptide bonds, and a preference for a large uncharged residue in P1. Hydrolyses peptide amides
Other names: alcalase; alcalase 0.6L; alcalase 2.5L; ALK-enzyme; bacillopeptidase A; bacillopeptidase B; Bacillus subtilis alkaline proteinase bioprase; bioprase AL 15; bioprase APL 30; colistinase; (see also comments); subtilisin J; subtilisin S41; subtilisin Sendai; subtilisin GX; subtilisin E; subtilisin BL; genenase I; esperase; maxatase; alcalase; thermoase PC 10; protease XXVII; thermoase; superase; subtilisin DY; subtilopeptidase; SP 266; savinase 8.0L; savinase 4.0T; kazusase; protease VIII; opticlean; Bacillus subtilis alkaline proteinase; protin A 3L; savinase; savinase 16.0L; savinase 32.0 L EX; orientase 10B; protease S
Comments: Subtilisin is a serine endopeptidase, type example of peptidase family S8. It contains no cysteine residues (although these are found in homologous enzymes). Species variants include subtilisin BPN' (also subtilisin B, subtilopeptidase B, subtilopeptidase C, Nagarse, Nagarse proteinase, subtilisin Novo, bacterial proteinase Novo) and subtilisin Carlsberg (subtilisin A, subtilopeptidase A, alcalase Novo). Formerly EC 3.4.4.16 and included in EC 3.4.21.14. Similar enzymes are produced by various Bacillus subtilis strains and other Bacillus species [1,3]
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 9014-01-1
References
1. Ottesen, M. and Svendsen, I. The subtilisins. Methods Enzymol. 19 (1970) 199-215
2. Markland, F.S. and Smith, E.L. Subtilisins: primary structure, chemical and physical properties In: The Enzymes, 3rd edn., vol. 3, (Boyer, P.D., ed.), pp. 561-608 (1971) Academic Press, New York
3. Philipp, M. and Bender, M.L. Kinetics of subtilisin and thiolsubtilisin. Mol. Cell. Biochem. 51 (1983) 5-32. [PMID: 6221910]
4. Nedkov, P., Oberthür, W. and Braunitzer, G. Determination of the complete amino acid sequence of subtilisin DY and its comparison with the primary structures of the subtilisins BPN, Carlsberg and amylosacchariticus. Biol. Chem. Hoppe-Seyler 366 (1985) 421-430. [PMID: 3927935]
5. Ikemura, H., Takagi, H. and Inouye, M. Requirement of pro-sequence for the production of active subtilisin E in Escherichia coli. J. Biol. Chem. 262 (1987) 7859-7864. [PMID: 3108260]
6. Polgár, L. Structure and function of serine proteases. In New Comprehensive Biochemistry Vol. 16, Hydrolytic Enzymes (Neuberger, A. and Brocklehurst,K., eds), pp. 159-200 (1987) Elsevier, Amsterdam
[EC 3.4.21.62 created 1992 (EC 3.4.21.14 created 1961 as EC 3.4.4.16, transferred 1972 to EC 3.4.21.14, modified 1986, part incorporated 1992)]
EC 3.4.21.63
Accepted name: oryzin
Reaction: Hydrolysis of proteins with broad specificity, and of Bz-Arg-OEt > Ac-Tyr-OEt. Does not hydrolyze peptide amides
Other names: Aspergillus alkaline proteinase; aspergillopeptidase B; API 21; aspergillopepsin B; aspergillopepsin F; Aspergillus candidus alkaline proteinase; Aspergillus flavus alkaline proteinase; Aspergillus melleus semi-alkaline proteinase; Aspergillus oryzae alkaline proteinase; Aspergillus parasiticus alkaline proteinase; Aspergillus serine proteinase; Aspergillus sydowi alkaline proteinase; Aspergillus soya alkaline proteinase; Aspergillus melleus alkaline proteinase; Aspergillus sulphureus alkaline proteinase; prozyme; P 5380; kyorinase; seaprose S; semi-alkaline protease; sumizyme MP; prozyme 10; onoprose; onoprose SA; protease P; promelase
Comments: A peptidase of family S8 (subtilisin family), not containing cysteine, that is the predominant extracellular alkaline endopeptidase of the mold Aspergillus oryzae. Formerly EC 3.4.21.15, and included in EC 3.4.21.14. Identical or closely related enzymes are produced by A. flavus and A. sojae [2,3,4]
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 9074-07-1
References
1. Nakagawa, Y. Alkaline proteinases from Aspergillus. Methods Enzymol. 19 (1970) 581-591
2. Hayashi, K. and Terada, M. Some characteristics of hydrolysis of synthetic substrates and proteins by the alkaline proteases from Aspergillus sojae. Agric. Biol. Chem. 36 (1972) 1755-1765
3. Turková, J., Mikes, O., Hayashi, K., Danno, G. and Polgár, L. Alkaline proteinases of the genus Aspergillus. Biochim. Biophys. Acta 257 (1972) 257-263. [PMID: 4623338]
4. Morihara, K., Oka, T. and Tsuzuki, H. Comparative study of various serine alkaline proteinases from microorganisms. Esterase activity against N-acylated peptide ester substrates. Arch. Biochem. Biophys. 165 (1974) 72-79. [PMID: 4441086]
5. Spadari, S., Subramanian, A.R. and Kalnitsky, G. Highly restricted specificity of the serine proteinase aspergillopeptidase B. Biochim. Biophys. Acta 359 (1974) 267-272. [PMID: 4859351]
[EC 3.4.21.63 created 1992 (EC 3.4.21.14 created 1961 as EC 3.4.4.16, transferred 1972 to EC 3.4.21.14, modified 1986, part incorporated 1992)]
EC 3.4.21.64
Accepted name: peptidase K
Reaction: Hydrolysis of keratin, and of other proteins with subtilisin-like specificity. Hydrolyses peptide amides
Other names: Tritirachium alkaline proteinase; Tritirachium album serine proteinase; proteinase K; Tritirachium album proteinase K; endopeptidase K
Comments: From the mold Tritirachium album Limber. A peptidase of family S8 (subtilisin family) containing two disulfide bridges and one free Cys near the active site His. Formerly included in EC 3.4.21.14
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 39450-01-6
References
1. Ebeling, W., Hennrich, N., Klockow, M., Metz, H., Orth, H.D. and Lang, H. Proteinase K from Tritirachium album Limber. Eur. J. Biochem. 47 (1974) 91-97. [PMID: 4373242]
2. Morihara, K. and Tsuzuki, H. Specificity of proteinase K from Tritirachium album Limber for synthetic peptides. Agric. Biol. Chem. 39 (1975) 1489-1492
3. Kraus, E., Klitz, H.H. and Fembert, U.F. The specificity of proteinase K against oxidized insulin B chain. Hoppe-Seyler's Z. Physiol. Chem. 357 (1976) 233-237. [PMID: 943367]
4. Jany, K.-D., Lederer, G. and Mayer, B. Amino acid sequence of proteinase K from the mold Tritirachium album Limber. FEBS Lett. 199 (1986) 139-144
5. Betzel, C., Teplyakov, A.V., Harutyunyan, E.H., Saenger, W. and Wilson, K.S. Thermitase and proteinase K: a comparison of the refined three-dimensional structures of the native enzymes. Protein Engineering 3 (1990) 161-172 [PMID: 2184432]
[EC 3.4.21.64 created 1992 (EC 3.4.21.14 created 1961 as EC 3.4.4.16, transferred 1972 to EC 3.4.21.14, modified 1986, part incorporated 1992)]
EC 3.4.21.65
Accepted name: thermomycolin
Reaction: Rather nonspecific hydrolysis of proteins. Preferential cleavage: Ala , Tyr
, Tyr , Phe
, Phe in small molecule substrates
in small molecule substrates
Other names: thermomycolase
Comments: A peptidase of family S8 (subtilisin family) from the thermophilic fungus Malbranchea pulchella var. sulfurea containing Cys, but not inhibited by p-mercuribenzoate. Very thermostable. Formerly included in EC 3.4.21.14
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number: 52233-31-5
References
1. Gaucher, G.M. and Stevenson, K.J. Thermomycolin. Methods Enzymol. 45 (1976) 415-433. [PMID: 1012007]
[EC 3.4.21.65 created 1992 (EC 3.4.21.14 created 1961 as EC 3.4.4.16, transferred 1972 to EC 3.4.21.14, modified 1986, part incorporated 1992)]
EC 3.4.21.66
Accepted name: thermitase
Reaction: Hydrolysis of proteins, including collagen
Other names: thermophilic Streptomyces serine proteinase; Thermoactinomyces vulgaris serine proteinase
Comments: A peptidase of family S8 (subtilisin family) from Thermoactinomyces vulgaris containing a single Cys, near the active site His, and inhibited by p-mercuribenzoate. The N-terminal extension of the polypeptide chain relative to subtilisin contributes to Ca2+-binding and the high thermostability. The amino acid composition and properties of the thermostable enzyme from Streptomyces rectus var. proteolyticus (formerly included in EC 3.4.21.14) are closely similar [1,2]
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 69772-87-8
References
1. Mizusawa, K. and Yoshida, F. Thermophilic Streptomyces alkaline proteinase. J. Biol. Chem. 247 (1972) 6978-6984. [PMID: 4711613]
2. Borgia, P. and Campbell, L. Properties of two homologous alkaline proteases from Streptomyces rectus. J. Bacteriol. 123 (1974) 1109-1115. [PMID: 4373436]
3. Kleine, R. Properties of thermitase, a thermostable serine protease from Thermoactinomyces vulgaris. Acta Biol. Med. Ger. 41 (1982) 89-102. [PMID: 7051706]
4. Meloun, B., Baudy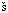 , M., Kostka, V., Hausdorf, G., Frömmel, C. and Höhne, W.E. Complete primary structure of thermitase from Thermoactinomyces vulgaris and its structural features related to the subtilisin-type proteinases. FEBS Lett. 183 (1985) 195-200
, M., Kostka, V., Hausdorf, G., Frömmel, C. and Höhne, W.E. Complete primary structure of thermitase from Thermoactinomyces vulgaris and its structural features related to the subtilisin-type proteinases. FEBS Lett. 183 (1985) 195-200
5. Teplyakov, A.V., Kuranova, I.P., Harutyunyan, E.H., Vainshtein, B.K., Frömmel, C., Höhne, W.E. and Wilson, K.S. Crystal structure of thermitase at 1.4 Å resolution. J. Mol. Biol. 214 (1990) 261-279. [PMID: 2196375]
[EC 3.4.21.66 created 1992]
EC 3.4.21.67
Accepted name: endopeptidase So
Reaction: Hydrolysis of proteins, but not Bz-Tyr-OEt, Ac-Phe-β-naphthylester, or Bz-Arg-OEt
Other names: E. coli cytoplasmic proteinase; proteinase So; Escherichia coli serine proteinase So
Comments: An Escherichia coli cytoplasmic endopeptidase formerly included in EC 3.4.21.14. Inhibited by Tos-Phe-CH2Cl, but not by Tos-Lys-CH2Cl
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number: 81611-83-8
References
1. Goldberg, A.L., Swamy, K.H.S., Chung, C.H. and Larimore, F.S. Proteases in Escherichia coli. Methods Enzymol. 80 (1981) 680-702.
2. Chung, C.H. and Goldberg, A.L. Purification and characterization of protease So, a cytoplasmic serine protease in Escherichia coli. J. Bacteriol. 154 (1983) 231-238. [PMID: 6339474]
[EC 3.4.21.67 created 1992 (EC 3.4.21.14 created 1961 as EC 3.4.4.16, transferred 1972 to EC 3.4.21.14, modified 1986, part incorporated 1992)]
EC 3.4.21.68
Accepted name: t-plasminogen activator
Reaction: Specific cleavage of Arg Val bond in plasminogen to form plasmin
Val bond in plasminogen to form plasmin
Other names: tissue plasminogen activator; plasminogen activator, tissue-type; tissue-type plasminogen activator; tPA; t-PA
Comments: A peptidase of family S1 (trypsin family) from a wide variety of mammalian tissues, especially endothelial cells. Secreted as a single chain precursor which is cleaved to a two-chain form by plasmin. Activity is considerably enhanced by fibrin. Formerly included in EC 3.4.21.31 and EC 3.4.99.26
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 139639-23-9
References
1. Pennica, D., Holmes, W.E., Kohr, W.J., Harkins, R.N., Vehar, G.A., Ward, C.A., Bennett, W.F., Yelverton, E., Seeburg, P.H., Heyneker, H.L., Goeddel, D.V. and Collen, D. Cloning and expression of human tissue-type plasminogen activator cDNA in E. coli. Nature 301 (1983) 214-221. [PMID: 6337343]
2. Loskutoff, D.J. and Schleef, R.R. Plasminogen activators and their inhibitors. Methods Enzymol. 163 (1988) 293-302. [PMID: 3148826]
3. Petersen, L.C., Johannessen, M., Forster, D., Kumar, A. and Mulvihill, E. The effect of polymerised fibrin on the catalytic activities of one-chain tissue-type plasminogen activator as revealed by an analogue resistant to plasmin cleavage. Biochim. Biophys. Acta 952 (1988) 245-254 [PMID: 2962643]
4. Verheijen, J.H. Tissue-type plasminogen activator and fast-acting plasminogen activator inhibitor in plasma. Methods Enzymol. 163 (1988) 302-309. [PMID: 3148827]
5. Gerard, R.D. and Meidell, R.S. Regulation of tissue plasminogen activator expression. Annu. Rev. Physiol. 51 (1989) 245-262. [PMID: 2496643]
6. Collen, D., Lijnen, H. R. and Verstraete, M. The fibrinolytic system and its disorders. In Blood: Principles and Practice of Hematology, 2nd edn (Handin, R.I., Lux, S.E. and Stossel, J.P., eds), J.B.Lippincott Company, Philadelphia (1990)
[EC 3.4.21.68 created 1972 as EC 3.4.99.26, transferred 1978 as EC 3.4.21.31, part transferred 1992 to EC 3.4.21.68]
EC 3.4.21.69
Accepted name: activated protein C (thrombin-activated peptidase)
Reaction: Degradation of blood coagulation factors Va and VIIIa
Other name(s): blood-coagulation factor XIVa; activated blood coagulation factor XIV; activated protein C; autoprothrombin II-A; protein Ca; APC; GSAPC; protein C (activated)
Comments: A peptidase of family S1 (trypsin family), one of the γ-carboxyglutamic acid-containing coagulation factors. The enzyme plays an important role in regulating anticoagulation, inflammation, and cell death by proteolytically inactivating the blood coagulation factors factor Va and factor VIIIa. Formed from protein C, the proenzyme that circulates in plasma, by the action of a complex of thrombin with thrombomodulin. Protein C can also be activated by serine endopeptidases present in several snake venoms.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc,
MEROPS,
PDB,
CAS registry number: 42617-41-4
References:
1. Esmon, C.T. The regulation of natural anticoagulant pathways. Science 235 (1987) 1348-1352. [PMID: 3029867]
2. Esmon, C.T. The roles of protein C and thrombomodulin in the regulation of blood coagulation. J. Biol. Chem. 264 (1989) 4743-4746. [PMID: 2538457]
[EC 3.4.21.69 created 1992, modified 2025]
EC 3.4.21.70
Accepted name: pancreatic endopeptidase E
Reaction: Preferential cleavage: Ala . Does not hydrolyse elastin
. Does not hydrolyse elastin
Other names: cholesterol-binding proteinase; proteinase E; cholesterol-binding serine proteinase; pancreatic protease E; pancreatic proteinase E; cholesterol-binding pancreatic proteinase; CBPP; pancreas E proteinase
Comments: A peptidase of family S1 (trypsin family) from pancreatic juice. Unlike elastases, has an acidic pI. Binds cholesterol
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number: 68073-27-8
References
1. Mallory, P.A. and Travis, J. Human pancreatic enzymes: purification and characterization of a nonelastolytic enzyme, protease E, resembling elastase. Biochemistry 14 (1975) 722-729. [PMID: 234742]
2. Shen, W., Fletcher, T.S. and Largman, C. Primary structure of human pancreatic protease E determined by sequence analysis of the cloned mRNA. Biochemistry 26 (1987) 3447-3452. [PMID: 3477287]
[EC 3.4.21.70 created 1992]
EC 3.4.21.71
Accepted name: pancreatic elastase II
Reaction: Preferential cleavage: Leu , Met
, Met and Phe
and Phe . Hydrolyses elastin
. Hydrolyses elastin
Other names: pancreatic elastase 2
Comments: A peptidase of family S1 (trypsin family) formed by activation of proelastase II from mammalian pancreas by trypsin. Usually, only one of the pancreatic elastases (see also EC 3.4.21.36) is expressed in a given species; human pancreatic elastase is of type II
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 75603-19-9
References
1. Fletcher, T.S., Shen, W.-F. and Largman, C. Primary structure of human pancreatic elastase 2 determined by sequence analysis of the cloned mRNA. Biochemistry 26 (1987) 7256-7261. [PMID: 3427074]
2. Shirasu, Y., Yoshida, H., Matsuki, S., Takemura, K., Ikeda, N., Shimada, Y., Ozawa, T., Mikayama, T., Iijima, H., Ishida, A., Sato, Y., Tamai, Y., Tanaka, J. and Ikenaga, H. Molecular cloning and expression in Escherichia coli of a cDNA encoding human pancreatic elastase 2. J. Biochem. (Tokyo) 102 (1987) 1555-1563. [PMID: 2834346]
[EC 3.4.21.71 created 1992]
EC 3.4.21.72
Accepted name: IgA-specific serine endopeptidase
Reaction: Cleavage of immunoglobulin A molecules at certain Pro bonds in the hinge region. No small molecule substrates are known
bonds in the hinge region. No small molecule substrates are known
Other names: IgA protease; IgA proteinase; IgA-specific proteinase; immunoglobulin A protease; immunoglobulin A proteinase
Comments: Species variants differing slightly in specificity are secreted by Gram-negative bacteria Neisseria gonorrhoeae and Haemophilus influenzae. Type example of peptidase family S6. Some other bacterial endopeptidases with similar specificity are of metallo- type (see EC 3.4.24.13, IgA-specific metalloendopeptidase)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 55127-02-1
References
1. Plaut, A.G. The IgA1 proteases of pathogenic bacteria. Annu. Rev. Microbiol. 37 (1983) 603-622. [PMID: 6416146]
2. Bachovchin, W.W., Plaut, A.G., Flentke, G.R., Lynch, M. and Kettner, C.A. Inhibition of IgA1 proteinases from Neisseria gonorrhoeae and Hemophilus influenzae by peptide prolyl boronic acids. J. Biol. Chem. 265 (1990) 3738-3743. [PMID: 2105953]
[EC 3.4.21.72 created 1992]
EC 3.4.21.73
Accepted name: u-plasminogen activator
Reaction: Specific cleavage of Arg Val bond in plasminogen to form plasmin
Val bond in plasminogen to form plasmin
Other names: urokinase; urinary plasminogen activator; cellular plasminogen activator; urokinase-type plasminogen activator; double-chain urokinase-type plasminogen activator; two-chain urokinase-type plasminogen activator; urokinase plasminogen activator; uPA; u-PA; abbokinase; urinary esterase A
Comments: Formed from the inactive precursor by action of plasmin or plasma kallikrein. Differs in structure from t-plasminogen activator (EC 3.4.21.68), and does not bind to fibrin. In peptidase family S1 (trypsin family). Formerly included in EC 3.4.21.31 and EC 3.4.99.26
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 9039-53-6
References
1. Lottenberg, R., Christiansen, U., Jackson, C.M. and Coleman, P.L. Assay of coagulation proteases using peptide chromogenic and fluorogenic substrates. Methods Enzymol. 80 (1981) 341-361. [PMID: 6210826]
2. Loskutoff, D.J. and Schleef, R.R. Plasminogen activators and their inhibitors. Methods Enzymol. 163 (1988) 293-302. [PMID: 3148826]
3. Saksela, O. and Rifkin, D.B. Cell-associated plasminogen activation: regulation and physiological functions. Annu. Rev. Cell Biol. 4 (1988) 93-126. [PMID: 3143380]
4. Collen, D., Lijnen, H.R. and Verstraete, M. The fibrinolytic system and its disorders. In Blood: Principles and Practice of Hematology, 2nd edn (Handin, R.I., Lux, S.E. and Stossel, J.P., eds), J.B.Lippincott Company, Philadelphia (1990)
5. Lijnen, H.R., Van Hoef, B., Nelles, L. and Collen, D. Plasminogen activation with single-chain urokinase-type plasminogen activator (scu-PA). Studies with active site mutagenized plasminogen (Ser740→Ala) and plasmin-resistant scu-PA (Lys158→Glu). J. Biol. Chem. 265 (1990) 5232-5236. [PMID: 1969415]
[EC 3.4.21.73 created 1972 as EC 3.4.99.26, transferred 1978 as EC 3.4.21.31, part transferred 1992 to EC 3.4.21.73]
EC 3.4.21.74
Accepted name: venombin A
Reaction: Selective cleavage of Arg bond in fibrinogen, to form fibrin, and release fibrinopeptide A. The specificity of further degradation of fibrinogen varies with species origin of the enzyme
bond in fibrinogen, to form fibrin, and release fibrinopeptide A. The specificity of further degradation of fibrinogen varies with species origin of the enzyme
Other names: α-fibrinogenase; habutobin; zinc metalloproteinase Cbfib1.1; zinc metalloproteinase Cbfib1.2; zinc metalloproteinase Cbfib2; ancrod; (see also Comments)
Comments: A somewhat thrombin-like enzyme from venoms of snakes of the viper/rattlesnake group. Species variants of the enzyme include ancrod from Agkistrodon rhodostoma (Malayan pit viper) (formerly EC 3.4.21.28) [1], batroxobin from Bothrops atrox (South American pit viper) (formerly EC 3.4.21.29) [2,5] and crotalase from Crotalus adamanteus (Eastern diamondback rattlesnake) (formerly EC 3.4.21.30) [3,4]. In peptidase family S1 (trypsin family). Does not require activation by Ca2+
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 146240-35-9
References
1. Nolan, C., Hall, L.S. and Barlow, G.H. Ancrod, the coagulating enzyme from Malayan pit viper (Agkistrodon rhodostoma) venom. Methods Enzymol. 45 (1976) 205-213. [PMID: 1011992]
2. Stocker, K. and Barlow, G.H. The coagulant enzyme from Bothrops atrox venom (batroxobin). Methods Enzymol. 45 (1976) 214-223. [PMID: 1011993]
3. Markland, F.S., Kettner, C., Schiffmann, S., Shaw, E., Bajwa, S.S., Reddy, K.N.N., Kirakossian, H., Patkos, G.B., Theodor, I. and Pirkle, H. Kallikrein-like activity of crotalase, a snake venom enzyme that clots fibrinogen. Proc. Natl. Acad. Sci. USA 79 (1982) 1688-1692. [PMID: 7043462]
4. Simmons, G., Bundalian, M., Theodor, I., Martinoli, J. and Pirkle, H. Action of crotalase, an enzyme with thrombin-like and kallikrein-like specificities, on tripeptide nitroanilide derivatives. Thromb. Res. 40 (1985) 555-561. [PMID: 2934864]
5. Itoh, N., Tanaka, N., Funakoshi, I., Kawasaki, T., Mihashi, S. and Yamashina, I. Organisation of the gene for batroxobin, a thrombin-like snake venom enzyme. Homology with the trypsin/kallikrein gene family. J. Biol. Chem. 263 (1988) 7628-7631. [PMID: 3163691]
[EC 3.4.21.74 created 1992 (EC 3.4.21.28, EC 3.4.21.29 and 3.4.21.30 all created 1978 and incorporated 1992)]
EC 3.4.21.75
Accepted name: furin
Reaction: Release of mature proteins from their proproteins by cleavage of -Arg-Xaa-Yaa-Arg bonds, where Xaa can by any amino acid and Yaa is Arg or Lys. Releases albumin, complement component C3 and von Willebrand factor from their respective precursors
bonds, where Xaa can by any amino acid and Yaa is Arg or Lys. Releases albumin, complement component C3 and von Willebrand factor from their respective precursors
Other names: prohormone convertase; dibasic processing enzyme; PACE; paired basic amino acid cleaving enzyme; paired basic amino acid converting enzyme; serine proteinase PACE; PC1; SPC3; proprotein convertase
Comments: One of a group of peptidases In peptidase family S8 (subtilisin family) that is structurally and functionally similar to kexin. All are activated by Ca2+, contain Cys near the active site His, and are inhibited by p-mercuribenzoate. At least three related enzymes are recognized in mammals: PC2, PC3 and PC4, which have somewhat different specificities
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 141760-45-4
References
1. Van de Ven, W.J.M., Voorberg, J., Fontijn, R., Pannekoek, H., van den Ouweland, A.M.W., van Duijnhoven, H.L.P., Roebroek, A.J.M. and Siezen, R.J. Furin is a subtilisin-like proprotein processing enzyme in higher eukaryotes. Mol. Biol. Rep. 14 (1990) 265-275. [PMID: 2094803]
2. Van de Ven, W.J.M., Creemers, J.W.M. and Roebroek, A.J.M. Furin: the prototype mammalian subtilisin-like proprotein-processing enzyme. Endoproteolytic cleavage at paired basic residues of proproteins of the eukaryotic secretory pathway. Enzyme 45 (1991) 257-270. [PMID: 1843280]
3. Hatsuzawa, K., Murakami, K. and Nakayama, K. Molecular and enzymatic properties of furin, a Kex2-like endoprotease involved in precursor cleavage at Arg-X-Lys/Arg-Arg sites. J. Biochem. (Tokyo) 111 (1992) 296-301. [PMID: 1587790]
4. Seidah, N.G. and Chrétien, M. Proprotein and prohormone convertases of the subtilisin family: recent developments and future perspectives. Trends Endocrinol. Metab. 3 (1992) 133-140
5. Steiner, D.F., Smeekens, S.P., Ohagi, S. and Chan, S.J. The new enzymology of precursor processing endoproteases. J. Biol. Chem. 267 (1992) 23435-23438. [PMID: 1429684]
[EC 3.4.21.75 created 1993]
EC 3.4.21.76
Accepted name: myeloblastin
Reaction: Hydrolysis of proteins, including elastin, by preferential cleavage: -Ala > -Val
> -Val
Other names: leukocyte proteinase 3; leukocyte proteinase 4; Wegener's granulomatosis autoantigen;proteinase PR-3; proteinase-3; PMNL proteinase
Comments: From polymorphonuclear leukocyte granules. In peptidase family S1 (trypsin family). Not inhibited by secretory leukocyte proteinase inhibitor
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 128028-50-2
References
1. Labbaye, C., Musette, P. and Cayre, Y.E. Wegener autoantigen and myeloblastin are encoded by a single mRNA. Proc. Natl. Acad. Sci. USA 88 (1991) 9253-9256. [PMID: 1681549]
2. Rao, N.V., Wehner, N.G., Marshall, B.C., Gray, W.R., Gray, B.H. and Hoidal, J.R. Characterization of proteinase-3 (PR-3), a neutrophil serine proteinase. Structural and functional properties. J. Biol. Chem. 266 (1991) 9540-9548. [PMID: 2033050]
3. Brubaker, M.J., Groutas, W.C., Hoidal, J.R. and Rao, N.V. Human neutrophil proteinase 3: mapping of the substrate binding site using peptidyl thiobenzyl esters. Biochem. Biophys. Res. Commun. 188 (1992) 1318-1324 [PMID: 1445363]
4. Kam, C.-M., Kerrigan, J.E., Dolman, K.M., Goldschmeding, R., von dem Borne, A.E.G.K. and Powers, J.C. Substrate and inhibitor studies on proteinase 3. FEBS Lett. 297 (1992) 119-123. [PMID: 1551417]
[EC 3.4.21.76 created 1993]
EC 3.4.21.77
Accepted name: semenogelase
Reaction: Preferential cleavage: -Tyr
Other names: prostate-specific antigen; α-seminoprotein; seminin; P-30 antigen; antigen (human clone λHPSA-1 prostate-specific protein moiety reduced); γ-seminoglycoprotein (human protein moiety reduced); γ-SM; antigen PSA (human prostate-specific); human glandular kallikrein; antigen PSA (human clone 5P1 protein moiety reduced)
Comments: A peptidase of family S1 (trypsin family) from seminal plasma. Slowly inhibited by α1-antichymotrypsin
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 110157-83-0 and 95829-41-7
References
1. Digby, M., Zhang, X.-Y. and Richards, R.I. Human prostate specific antigen (PSA) gene: structure and linkage to the kallikrein-like gene, hGK-1. Nucleic Acids Res. 15 (1989) 2137 only. [PMID: 2467258]
2. Christensson, A., Laurell, C.-B. and Lilja, H. Enzymatic activity of prostate-specific antigen and its reactions with extracellular serine proteinase inhibitors. Eur. J. Biochem. 194 (1990) 755-763. [PMID: 1702714]
[EC 3.4.21.77 created 1993]
EC 3.4.21.78
Accepted name: granzyme A
Reaction: Hydrolysis of proteins, including fibronectin, type IV collagen and nucleolin. Preferential cleavage: -Arg , -Lys
, -Lys >> -Phe
>> -Phe in small molecule substrates
in small molecule substrates
Other names: CTLA3; HuTPS; T-cell associated protease 1; cytotoxic T lymphocyte serine protease; TSP-1; T-cell derived serine proteinase
Comments: From cytotoxic T lymphocyte granules. In peptidase family S1 (trypsin family). The human enzyme does not cleave Phe -
-
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 143180-73-8
References
1. Simon, M.M., Hoschützky, H., Fruth, U., Simon, H.-G. and Kramer, M.D. Purification and characterization of a T cell specific serine proteinase (TSP-1) from cloned cytolytic T lymphocytes. EMBO J. 5 (1986) 3267-3274 [PMID: 3545816]
2. Gershenfeld, H.K., Hershberger, R.J., Shows, T.B. and Weissman, I.L. Cloning and chromosomal assignment of a human cDNA encoding a T cell- and natural killer cell-specific trypsin-like serine protease. Proc. Natl. Acad. Sci. USA 85 (1988) 1184-1188. [PMID: 3257574]
3. Odake, S., Kam, C.-M., Narasimhan, L., Poe, M., Blake, J.T., Krahenbuhl, O., Tschopp, J. and Powers, J.C. Human and murine cytotoxic T lymphocyte serine proteases: subsite mapping with peptide thioester substrates and inhibition of enzyme activity and cytolysis by isocoumarins. Biochemistry 30 (1991) 2217-2227. [PMID: 1998680]
[EC 3.4.21.78 created 1993]
EC 3.4.21.79
Accepted name: granzyme B
Reaction: Preferential cleavage: -Asp >> -Asn
>> -Asn > -Met
> -Met , -Ser
, -Ser
Other names: CTLA1; CCPII; cytotoxic cell proteinase-1; granzyme G; granzyme H; CCP1 proteinase
Comments: From cytotoxic T lymphocyte granules. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 143180-74-9
References
1. Schmid, J. and Weissmann, C. Induction of mRNA for a serine protease and a β-thromboglobulin-like protein in mitogen-stimulated human leukocytes. J. Immunol. 139 (1987) 250-256. [PMID: 2953813]
2. Odake, S., Kam, C.-M., Narasimhan, L., Poe, M., Blake, J.T., Krahenbuhl, O., Tschopp, J. and Powers, J.C. Human and murine cytotoxic T lymphocyte serine proteases: subsite mapping with peptide thioester substrates and inhibition of enzyme activity and cytolysis by isocoumarins. Biochemistry 30 (1991) 2217-2227. [PMID: 1998680]
3. Poe, M., Blake, J.T., Boulton, D.A., Gammon, M., Sigal, N.H., Wu, J.K. and Zweerink, H.J. Human cytotoxic lymphocyte granzyme B. Its purification from granules and the characterization of substrate and inhibitor specificity. J. Biol. Chem. 266 (1991) 98-103. [PMID: 1985927]
[EC 3.4.21.79 created 1993]
EC 3.4.21.80
Accepted name: streptogrisin A
Reaction: Hydrolysis of proteins with specificity similar to chymotrypsin
Other names: Streptomyces griseus protease A; protease A; proteinase A; Streptomyces griseus proteinase A; Streptomyces griseus serine proteinase 3; Streptomyces griseus serine proteinase A
Comments: From Streptomyces griseus. A component of Pronase, In family S1 (trypsin family). Not inhibited by Tos-Phe-CH2Cl or ovomucoid
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 55326-50-6
References
1. Johnson, P. and Smillie, L.B. The disulfide bridge sequences of a serine protease of wide specificity from Streptomyces griseus. Can. J. Biochem. 49 (1971) 548-562. [PMID: 5575653]
2. Sielecki, A.R., Hendrickson, W.A., Broughton, C.G., Delbaere, L.T.J., Brayer, G.D. and James, M.N.G. Protein structure refinement: Streptomyces griseus serine protease A at 1.8 Å resolution. J. Mol. Biol. 134 (1979) 781-804. [PMID: 119870]
3. James, M.N.G., Sielecki, A.R., Brayer, G.D., Delbaere, L.T.J. and Bauer, C.-A. Structures of product and inhibitor complexes of Streptomyces griseus protease A at 1.8 Å resolution. J. Mol. Biol. 144 (1980) 43-88. [PMID: 6783761]
4. Delbaere, L.T.J. and Brayer, G.D. The 1.8 Å structure of the complex between chymostatin and Streptomyces griseus protease A. A model for serine protease catalytic tetrahedral intermediates. J. Mol. Biol. 183 (1985) 89-103. [PMID: 3892018]
5. Henderson, G., Krygsman, P., Liu, C.J., Davey, C.C. and Malek, L.T. Characterization and structure of genes for proteases A and B from Streptomyces griseus. J. Bacteriol. 169 (1987) 3778-3784. [PMID: 3112129]
[EC 3.4.21.80 created 1993]
EC 3.4.21.81
Accepted name: streptogrisin B
Reaction: Hydrolysis of proteins with trypsin-like specificity
Other names: Streptomyces griseus protease B; pronase B; serine proteinase B; Streptomyces griseus proteinase B; Streptomyces griseus proteinase 1; Streptomyces griseus serine proteinase B
Comments: From Streptomyces griseus. A component of Pronase, In peptidase family S1 (trypsin family), distinct from Streptomyces trypsin
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 55071-87-9
References
1. Jurasek, L., Fackre, D. and Smillie, L.B. Remarkable homology about the disulfide bridges of a trypsin-like enzyme from Streptomyces griseus. Biochem. Biophys. Res. Commun. 37 (1969) 99-105. [PMID: 4899581]
2. Fujinaga, M., Read, R.J., Sielecki, A., Ardelt, W., Laskowski, M., Jr and James, M.N.G. Refined crystal structure of the molecular complex of Streptomyces griseus protease B, a serine protease, with the third domain of the ovomucoid inhibitor from turkey. Proc. Natl. Acad. Sci. USA 79 (1982) 4868-4872. [PMID: 6750612]
3. Read, R.J., Fujinaga, M., Sielecki, A.R. and James, M.N.G. Structure of the complex of Streptomyces griseus protease B and the third domain of turkey ovomucoid inhibitor at 1.8-Å resolution. Biochemistry 22 (1983) 4420-4433. [PMID: 6414511]
4. Henderson, G., Krygsman, P., Liu, C.J., Davey, C.C. and Malek, L.T. Characterization and structure of genes for proteases A and B from Streptomyces griseus. J. Bacteriol. 169 (1987) 3778-3784 [PMID: 3112129]
5. Greenblatt, H.M., Ryan, C.A. and James, M.N.G. Structure of the complex of Streptomyces griseus proteinase B and polypeptide chymotrypsin inhibitor-1 from Russet Burbank potato tubers at 2.1 Å resolution. J. Mol. Biol. 205 (1989) 201-228. [PMID: 2494344]
[EC 3.4.21.81 created 1993]
EC 3.4.21.82
Accepted name: glutamyl endopeptidase II
Reaction: Preferential cleavage: -Glu >> -Asp
>> -Asp . Preference for Pro or Leu at P2 and Phe at P3. Cleavage of -Glu
. Preference for Pro or Leu at P2 and Phe at P3. Cleavage of -Glu Asp- and -Glu
Asp- and -Glu Pro- bonds is slow
Pro- bonds is slow
Other names: GluSGP
Comments: From Streptomyces griseus. A peptidase of family S1 (trypsin family). Inhibited by [Leu18→Glu]-modified turkey ovomucoid third domain
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 137010-42-5
References
1. Yoshida, N., Tsuruyama, S., Nagata, K., Hirayama, K., Noda, K. and Makisumi, S. Purification and characterization of an acidic amino acid specific endopeptidase of Streptomyces griseus obtained from a commercial preparation (Pronase). J. Biochem. (Tokyo) 104 (1988) 451-456. [PMID: 3149277]
2. Komiyama, T., Bigler, T.L., Yoshida, N., Noda, K. and Laskowski, M., Jr Replacement of P1 Leu18 by Glu18 in the reactive site of turkey ovomucoid third domain converts it into a strong inhibitor of Glu-specific Streptomyces griseus Proteinase (GluSGP). J. Biol. Chem. 266 (1991) 10727-10730. [PMID: 1674942]
3. Nagata, K., Yoshida, N., Ogata, F., Araki, M. and Noda, K. Subsite mapping of an acidic amino acid-specific endopeptidase from Streptomyces griseus, GluSGP, and protease V8. J. Biochem. (Tokyo) 110 (1991) 859-862. [PMID: 1794975]
4. Svendsen, I., Jensen, M.R. and Breddam, K. The primary structure of the glutamic acid-specific protease of Streptomyces griseus. FEBS Lett. 292 (1991) 165-167. [PMID: 1959600]
5. Breddam, K. and Meldal, M. Substrate preferences of glutamic-acid-specific endopeptidases assessed by synthetic peptide substrates based on intramolecular fluorescence quenching. Eur. J. Biochem. 206 (1992) 103-107. [PMID: 1587264]
[EC 3.4.21.82 created 1993]
EC 3.4.21.83
Accepted name: oligopeptidase B
Reaction: Hydrolysis of -Arg , -Lys
, -Lys bonds in oligopeptides, even when P1' residue is proline
bonds in oligopeptides, even when P1' residue is proline
Other names: protease II; Escherichia coli alkaline proteinase II; protease II
Comments: Known from Escherichia coli. Inhibited by Tos-Lys-CH2Cl. In peptidase family S9 (prolyl oligopeptidase family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 57657-67-7
References
1. Kanatani, A., Masuda, T., Shimoda, T., Misoka, F., Lin, X.S., Yoshimoto, T. and Tsuru, D. Protease II from Escherichia coli: sequencing and expression of the enzyme gene and characterization of the expressed enzyme. J. Biochem. (Tokyo) 110 (1991) 315-320. [PMID: 1769955]
[EC 3.4.21.83 created 1993]
EC 3.4.21.84
Accepted name: limulus clotting factor 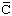
Reaction: Selective cleavage of -Arg103 Ser- and -Ile124
Ser- and -Ile124 Ile- bonds in limulus clotting factor B to form factor
Ile- bonds in limulus clotting factor B to form factor 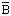 . Cleavage of -Pro-Arg
. Cleavage of -Pro-Arg bonds in synthetic substrates
bonds in synthetic substrates
Other name(s): factor C; limulus factor C
Comments: From the hemocyte granules of the horseshoe crabs Limulus and Tachypleus. Factor C is activated by Gram-negative bacterial lipopolysaccharides and chymotrypsin. Inhibited by antithrombin III. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number: 115743-27-6
References
1. Nakamura, T., Morita, T. and Iwanaga, S. Lipopolysaccharide-sensitive serine-protease zymogen (factor C) found in Limulus hemocytes. Isolation and characterization. Eur. J. Biochem. 154 (1986) 511-521. [PMID: 3512266]
2. Muta, T., Miyata, T., Misumi, Y., Tokunaga, F., Nakamura, T., Toh, Y., Ikehara, Y. and Iwanaga, S. Limulus factor C. An endotoxin-sensitive serine protease zymogen with a mosaic structure of complement-like, epidermal growth factor-like, and lectin-like domains. J. Biol. Chem. 266 (1991) 6554-6561. [PMID: 2007602]
3. Tokunaga, F., Nakajima, H. and Iwanaga, S. Further studies on lipopolysaccharide-sensitive serine protease zymogen (factor C): its isolation from Limulus polyphemus hemocytes and identification as an intracellular zymogen activated by α-chymotrypsin, not by trypsin. J. Biochem. (Tokyo) 109 (1991) 150-157. [PMID: 2016264]
[EC 3.4.21.84 created 1993]
EC 3.4.21.85
Accepted name: limulus clotting factor 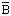
Reaction: Selective cleavage of -Arg98 Ile- bond in limulus proclotting enzyme to form active clotting enzyme
Ile- bond in limulus proclotting enzyme to form active clotting enzyme
Comments: From the hemocyte granules of the horseshoe crabs Limulus and Tachypleus. Factor B is activated by limulus clotting factor 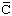 . In peptidase family S1 (trypsin family)
. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number: 848851-53-6
References
1. Nakamura, T., Horiuchi, T., Morita, T. and Iwanaga, S. Purification and properties of intracellular clotting factor, factor B, from horseshoe crab (Tachypleus tridentatus) hemocytes. J. Biochem. (Tokyo) 99 (1986) 847-857. [PMID: 3519594]
[EC 3.4.21.85 created 1993]
EC 3.4.21.86
Accepted name: limulus clotting enzyme
Reaction: Selective cleavage of -Arg18 and -Arg47
and -Arg47 bonds in coagulogen to form coagulin and fragments
bonds in coagulogen to form coagulin and fragments
Other name(s): clotting enzyme
Comments: From the hemocyte granules of horseshoe crabs Limulus and Tachypleus. Proclotting enzyme is activated by limulus clotting factor . In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number:
References
1. Muta, T., Hashimoto, R., Miyata, T., Nishimura, H., Toh, Y. and Iwanaga, S. Proclotting enzyme from horseshoe crab hemocytes. cDNA cloning, disulfide locations, and subcellular localization. J. Biol. Chem. 265 (1990) 22426-22433. [PMID: 2266134]
2. Tokunaga, F., Nakajima, H. and Iwanaga, S. Further studies on lipopolysaccharide-sensitive serine protease zymogen (factor C): its isolation from Limulus polyphemus hemocytes and identification as an intracellular zymogen activated by α-chymotrypsin, not by trypsin. J. Biochem. (Tokyo) 109 (1991) 150-157. [PMID: 2016264]
[EC 3.4.21.86 created 1993]
[EC 3.4.21.87 Transferred entry: now EC 3.4.23.49, omptin. The enzyme is not a serine protease, as thought previously, but an aspartate protease. (EC 3.4.21.87 created 1993, deleted 2006)]
EC 3.4.21.88
Accepted name: repressor LexA
Reaction: Hydrolysis of Ala84 Gly bond in repressor LexA
Gly bond in repressor LexA
Other names: LexA repressor
Comments: RecA protein and single-stranded DNA are required for activity, which is attributed to a Ser/Lys dyad [2]. The LexA protein represses the SOS regulon, which regulates the genes involved in DNA repair. In the presence of single-stranded DNA, the RecA protein interacts with repressor LexA, causing it to undergo an autocatalytic cleavage which disrupts the DNA-binding part of the repressor, and inactivates it. The consequent derepression of the SOS regulon leads to DNA repair. This peptidase activity of LexA was previously attributed to the RecA protein (formerly EC 3.4.99.37). Type example of peptidase family S24
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 84721-00-6
References
1. Horii, T., Ogawa, T. and Ogawa, H. Nucleotide sequence of the LexA gene of E. coli. Cell 23 (1981) 689-697. [PMID: 7013987]
2. Slilaty, S.N. and Little, J.W. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc. Natl. Acad. Sci. USA 84 (1987) 3987-3991. [PMID: 3108885]
3. Kim, B. and Little, J.W. LexA and λ CI repressors as enzymes: specific cleavage in an intermolecular reaction. Cell 73 (1993) 1165-1173. [PMID: 8513500]
4. Little, J.W., Kim, B., Roland, K.L., Smith, M.H., Lin, L.-L. and Slilaty, S.N. Cleavage of LexA repressor. Methods Enzymol. 244 (1994) 266-284. [PMID: 7845214]
[EC 3.4.21.88 created 1995]
EC 3.4.21.89
Accepted name: signal peptidase I
Reaction: Cleavage of hydrophobic, N-terminal signal or leader sequences
Other names: leader peptidase I; signal proteinase; Escherichia coli leader peptidase; eukaryotic signal peptidase; eukaryotic signal proteinase; leader peptidase; leader peptide hydrolase; leader proteinase; signal peptidase; pilin leader peptidase; SPC; prokaryotic signal peptidase; prokaryotic leader peptidase; HOSP; prokaryotic signal proteinase; propeptidase; PuIO prepilin peptidase; signal peptide hydrolase; signal peptide peptidase; signalase; bacterial leader peptidase 1; pilin leader peptidase
Comments: The enzyme is found in bacterial membranes and in chloroplast thylakoid membranes. Unaffected by inhibitors of most serine peptidases, but site-directed mutagenesis implicates a Ser/Lys catalytic dyad in activity [1,3]. Hydrolyses a single bond -Ala Ala- in M13 phage procoat protein, producing free signal peptide and coat protein. Formerly included in EC 3.4.99.36. Eukaryote signal peptidases that may have somewhat different specificity are known from the endoplasmic reticulum membrane [4] and mitochondrial inner membrane [2]. Type example of peptidase family S26
Ala- in M13 phage procoat protein, producing free signal peptide and coat protein. Formerly included in EC 3.4.99.36. Eukaryote signal peptidases that may have somewhat different specificity are known from the endoplasmic reticulum membrane [4] and mitochondrial inner membrane [2]. Type example of peptidase family S26
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 65979-36-4
References
1. Black, M.T. Evidence that the catalytic activity of prokaryote leader peptidase depends upon the operation of a serine-lysine catalytic dyad. J. Bacteriol. 175 (1993) 4957-4961. [PMID: 8394311]
2. Nunnari, J., Fox, T.D. and Walter, P. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science 262 (1993) 1997-2004
3. Tschantz, W.R., Sung, M., Delgado-Partin, V.M. and Dalbey, R.E. A serine and a lysine residue implicated in the catalytic mechanism of the Escherichia coli leader peptidase. J. Biol. Chem. 268 (1993) 27349-27354. [PMID: 8262975]
4. Lively, M.O., Newsome, A.L. and Nusier, M. Eukaryote microsomal signal peptidases. Methods Enzymol. 244 (1994) 301-314. [PMID: 7845216]
5. Tschantz, W.R. and Dalbey, R.E. Bacterial leader peptidase I. Methods Enzymol. 244 (1994) 285-301. [PMID: 7845215]
6. Chaal, B.K., Mould, R.M., Barbrook, A.C., Gray, J.C. and Howe, C.J. Characterization of a cDNA encoding the thylakoidal processing peptidase from Arabidopsis thaliana. Implications for the origin and catalytic mechanism of the enzyme. J. Biol. Chem. 273 (1998) 689-692. [PMID: 9422718]
7. Inoue, K., Baldwin, A.J., Shipman, R.L., Matsui, K., Theg, S.M. and Ohme-Takagi, M. Complete maturation of the plastid protein translocation channel requires a type I signal peptidase. J. Cell Biol. 171 (2005) 425-430. [PMID: 16275749]
[EC 3.4.21.89 created 1984 as EC 3.4.99.36, transferred 1995 to EC 3.4.21.89]
EC 3.4.21.90
Accepted name: togavirin
Reaction: Autocatalytic release of the core protein from the N-terminus of the togavirus structural polyprotein by hydrolysis of a -Trp Ser- bond
Ser- bond
Other names: Sindbis virus protease; Sindbis virus core protein; NsP2 proteinase
Comments: Known from the Sindbis and Semliki forest togaviruses. Once released, the core protein does not retain catalytic activity. Togavirin is the type example of peptidase family S3 and has a similar tertiary structure to chymotrypsin [3]
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 342882-56-8
References
1. Kräusslich, H.-G. and Wimmer, E. Viral proteinases. Annu. Rev. Biochem. 57 (1988) 701-754. [PMID: 3052288]
2. Strauss, E.G., De Groot, R.J., Levinson, R. and Strauss, J.H. Identification of the active site residues in the nsP2 proteinase of Sindbis virus. Virology 191 (1992) 932-940. [PMID: 1448929]
3. Tong, L., Wengler, G. and Rossmann, M.G. Refined structure of Sindbis virus core protein and comparison with other chymotrypsin-like serine proteinase structures. J. Mol. Biol. 230 (1993) 228-247. [PMID: 8450538]
[EC 3.4.21.90 created 1995]
EC 3.4.21.91
Accepted name: flavivirin
Reaction: Selective hydrolysis of -Xaa-Xaa Yaa- bonds in which each of the Xaa can be either Arg or Lys and Yaa can be either Ser or Ala
Yaa- bonds in which each of the Xaa can be either Arg or Lys and Yaa can be either Ser or Ala
Other names: Yellow fever virus (flavivirus) protease; NS2B-3 proteinase
Comments: Known from classical flaviviruses (yellow fever, dengue fever). The functional viral peptidase is part of the NS2B protein. Catalytic His, Asp and Ser residues are arranged as in chymotrypsin, but flavivrin is the type example of peptidase family S7
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 154215-26-6
References
1. Chambers, T.J., Hahn, C.S., Galler, R. and Rice, C.M. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44 (1990) 649-688. [PMID: 2174669]
2. Cahour, A., Falgout, B. and Lai, C.-J. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J. Virol. 66 (1992) 1535-1542. [PMID: 1531368]
3. Lin, C., Amberg, S.M., Chambers, T.J. and Rice, C.M. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J. Virol. 67 (1993) 2327-2335. [PMID: 8445732]
[EC 3.4.21.91 created 1995]
EC 3.4.21.92
Accepted name: endopeptidase Clp
Reaction: Hydrolysis of proteins to small peptides in the presence of ATP and Mg2+. α-Casein is the usual test substrate. In the absence of ATP, only oligopeptides shorter than five residues are hydrolysed (such as succinyl-Leu-Tyr NHMec; and Leu-Tyr-Leu
NHMec; and Leu-Tyr-Leu Tyr-Trp, in which cleavage of the -Tyr
Tyr-Trp, in which cleavage of the -Tyr Leu- and -Tyr
Leu- and -Tyr Trp bonds also occurs)
Trp bonds also occurs)
Other names: endopeptidase Ti; caseinolytic protease; protease Ti; ATP-dependent Clp protease; endopeptidase Ti; caseinolytic protease; ClpP; Clp protease
Comments: An enzyme from bacteria that contains subunits of two types, ClpP, with peptidase activity, and ClpA, with ATPase activity. The ClpAP complex, which displays ATP-dependent endopeptidase activity, has the composition (ClpP14ClpA6)2 [4]. ClpP is the type example of peptidase family S14
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 110910-59-3
References
1. Gottesman, S., Clark, W.P. and Maurizi, M.R. The ATP-dependent Clp protease of Escherichia coli. Sequence of clpA and identification of a Clp-specific substrate. J. Biol. Chem. 265 (1990) 7886-7893 [PMID: 2186030]
2. Maurizi, M.R., Clark, W.P., Katayama, Y., Rudikoff, S., Pumphrey, J., Bowers, B. and Gottesman, S. Sequence and structure of Clp P, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J. Biol. Chem. 265 (1990) 12536-12545. [PMID: 2197275]
3. Maurizi, M.R., Thompson, M.W., Singh, S.K. and Kim, S.-H. Endopeptidase Clp: the ATP-dependent Clp protease from Escherichia coli. Methods Enzymol. 244 (1994) 314-331. [PMID: 7845217]
4. Kessel, M., Maurizi,M.R., Kim, B., Kocsis, E., Trus, B., Singh, S.K. and Steven, A.C. Homology in structural organization between E. coli ClpAP protease and the eukaryotic 26 S proteasome. J. Mol. Biol. 250 (1995) 587-594. [PMID: 7623377]
[EC 3.4.21.92 created 1996]
EC 3.4.21.93
Accepted name: proprotein convertase 1
Reaction: Release of protein hormones, neuropeptides and renin from their precursors, generally by hydrolysis of -Lys-Arg bonds
bonds
Other names: prohormone convertase 3; neuroendocrine convertase 1; PC1
Comments: A Ca2+-dependent enzyme, maximally active at about pH 5.5. Substrates include pro-opiomelanocortin, prorenin, proenkephalin, prodynorphin, prosomatostatin and proinsulin. Unlike prohormone convertase 2, does not hydrolyse proluteinizing-hormone-releasing-hormone. Unusually, processing of prodynorphin occurs at a bond in which P2 is Thr. Present in the regulated secretory pathway of neuroendocrine cells, commonly acting co-operatively with prohormone convertase 2. In peptidase family S8 (subtilisin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 99676-46-7
References
1. Seidah, N.G., Gaspar, L., Mion, P., Marcinkiewicz, M., Mbikay, M. and Chrétien, M. cDNA sequence of two distinct pituitary proteins homologous to Kex2 and furin gene products: tissue-specific mRNAs encoding candidates for pro-hormone processing proteinases. DNA Cell Biol. 9 (1990) 415-424. [PMID: 2169760]
2. Smeekens, S.P., Avruch, A.S., LaMendola, J., Chan, S.J. and Steiner, D.F. Identification of a cDNA encoding a second putative prohormone convertase related to PC2 in AtT20 cells and islets of Langerhans. Proc. Natl. Acad. Sci. USA 88 (1991) 340-344. [PMID: 1988934]
3. Steiner, D.F., Smeekens, S.P., Ohagi, S. and Chan, S.J. The new enzymology of precursor processing endoproteases. J. Biol. Chem. 267 (1992) 23435-23438. [PMID: 1429684]
4. Seidah, N.G. and Chrétien, M. Pro-protein convertases of the subtilisin/kexin family. Methods Enzymol. 244 (1994) 175-188 [PMID: 7845206]
5. Jean, F., Basak, A., Dimaio, J., Seidah, N.G. and Lazure, C. An internally quenched fluorogenic substrate of prohormone convertase 1 and furin leads to a potent prohormone convertase inhibitor. Biochem. J. 307 (1995) 689-695. [PMID: 7741698]
[EC 3.4.21.93 created 1996]
EC 3.4.21.94
Accepted name: proprotein convertase 2
Reaction: Release of protein hormones and neuropeptides from their precursors, generally by hydrolysis of -Lys-Arg bonds
bonds
Other names: neuroendocrine convertase 2; PC2
Comments: A Ca2+-dependent enzyme, maximally active at about pH 5.5. Specificity is broader than that of prohormone convertase 1. Substrates include pro-opiomelanocortin, proenkephalin, prodynorphin, proglucagon, proinsulin and proluteinizing-hormone-releasing-hormone. Does not hydrolyse prorenin or prosomatostatin, however. Unusually, processing of prodynorphin occurs at a bond in which P2 is Thr. Present in the regulated secretory pathway of neuroendocrine cells, commonly acting co-operatively with prohormone convertase 1. In peptidase family S8 (subtilisin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number: 388092-42-0
References
1. Seidah, N.G., Gaspar, L., Mion, P., Marcinkiewicz, M., Mbikay, M. and Chrétien, M. cDNA sequence of two distinct pituitary proteins homologous to Kex2 and furin gene products: tissue- specific mRNAs encoding candidates for pro-hormone processing proteinases. DNA Cell Biol. 9 (1990) 415-424. [PMID: 2169760]
2. Smeekens, S.P. and Steiner, D.F. Identification of a human insulinoma cDNA encoding a novel mammalian protein structurally related to the yeast dibasic processing protease Kex2. J. Biol. Chem. 265 (1990) 2997-3000 [PMID: 2154467]
3. Rouillé, Y., Westermark, G., Martin, S.K. and Steiner, D.F. Proglucagon is processed to glucagon by prohormone convertase PC2 in αTC1-6 cells. Proc. Natl. Acad. Sci. USA 91 (1994) 3242-3246. [PMID: 8159732]
4. Seidah, N.G. and Chrétien, M. Pro-protein convertases of the subtilisin/kexin family. Methods Enzymol. 244 (1994) 175-188 [PMID: 7845206]
[EC 3.4.21.94 created 1996]
EC 3.4.21.95
Accepted name: snake venom factor V activator
Reaction: Fully activates human clotting factor V by a single cleavage at the Trp-Tyr-Leu-Arg1545 Ser-Asn-Asn-Gly bond. Cattle, but not rabbit, factor V is cleaved, and no other proteins of the clotting system are attacked. Esterase activity is observed on Bz-Arg-OEt and Tos-Arg-OMe, and amidase activity on Phe-pipecolyl-Arg-NHPhNO2
Ser-Asn-Asn-Gly bond. Cattle, but not rabbit, factor V is cleaved, and no other proteins of the clotting system are attacked. Esterase activity is observed on Bz-Arg-OEt and Tos-Arg-OMe, and amidase activity on Phe-pipecolyl-Arg-NHPhNO2
Comment: Known from venom of Vipera russelli. Inhibited by di-isopropyl fluorophosphate, unlike the metallopeptidase russellysin (EC 3.4.24.58) that is specific for factor X [1]. In peptidase family S1 (trypsin family) [2]
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 471269-12-2, 123757-15-3, 123757-16-4, 123757-17-5 and 23757-18-6
References
1. Kisiel, W. and Canfield, W. M. Snake venom proteases that activate blood-coagulation factor V. Methods Enzymol. 80 (1981) 275-285. [PMID: 7043192]
2. Tokunaga, F., Nagasawa, K., Tamura, S., Miyata, T., Iwanaga, S. and Kisiel, W. The factor V-activating enzyme (RVV-V) from Russell's viper venom. Identification of isoproteins RVV-Vα, -Vβ and -Vγ and their complete amino acid sequences. J. Biol. Chem. 263 (1988) 17417-17481 [PMID: 3053712]
[EC 3.4.21.95 created 1997]
EC 3.4.21.96
Accepted name: lactocepin
Reaction: Endopeptidase activity with very broad specificity, although some subsite preferences have been noted, e.g. large hydrophobic residues in the P1 and P4 positions, and Pro in the P2 position [1,2]. Best known for its action on caseins, although it has been shown to hydrolyse hemoglobin and oxidized insulin B chain
Other names: CEP; extracellular lactococcal proteinase; lactococcal cell wall-associated proteinase; lactococcal cell envelope-associated proteinase; lactococcal proteinase; PrtP
Comments: associated with the cell envelope of Lactococcus lactis and attached via a C-terminal membrane anchor sequence. Responsible for the hydrolysis of casein in milk and the provision of peptides essential to cell growth. Important in cheese making and the production of lactic casein, being required for rapid growth to high cell densities with concomitant production of adequate levels of lactic acid. Specificity differences between lactocepins from different starter strains may be partly responsible for imparting different flavour qualities to cheese [4]. In peptidase family S8 (subtilisin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number: 205510-58-3
References
1. Visser, S., Robben, A.J.P.M. and Slangen, C.J. Specificity of a cell-envelope-located proteinase (PIII-type) from Lactococcus lactis subsp. cremoris AM1 in its action on bovine beta-casein. Appl. Microbiol. Biotechnol. 35 (1991) 477-483. [PMID: 1367552]
2. Monnet, V., Ley, J.P. and Gonzalez, S. Substrate specificity of the cell envelope-located proteinase of Lactococcus lactis subsp. lactis NCDO763. Int. J. Biochem. 24 (1992) 707-718. [PMID: 1592148]
3. Exterkate, F.A., Alting, A.C. and Bruinenberg, P.G. Diversity of cell envelope proteinase specificity among strains of Lactococcus lactis and its relationship to charge characteristics of the substrate-binding region. Appl. Environ. Microbiol. 59 (1993) 3640-3647. [PMID: 8285671]
4. Pritchard, G.G. and Coolbear, T. The physiology and biochemistry of the proteolytic system in lactic acid bacteria. FEMS Microbiol. Rev. 12 (1993) 179-206. [PMID: 8398214]
[EC 3.4.21.96 created 1997]
EC 3.4.21.97
Accepted name: assemblin
Reaction: Cleaves -Ala Ser- and -Ala
Ser- and -Ala Ala- bonds in the scaffold protein
Ala- bonds in the scaffold protein
Comments: Involved in the breakdown of the scaffold protein during the late stages of assembly of the herpes-virus virion. Inhibited by diisopropyl fluorophosphate. Type example of peptidase family S21. Catalytic residues are His, Ser, His, a combination not known for any other peptidase, and the protein fold also is unique. Known from herpes viruses of several types, cytomegalovirus, Epstein-Barr virus and human herpesvirus 3
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 139691-88-6
References
1. Chen, P., Tsuge, H., Almassy, R.J., Gribskov, C.L., Katoh, S., Vanderpool, D.L., Margosiak, S.A., Pinko, C., Matthews, D.A. and Kan, C.C. Structure of the human cytomegalovirus protease catalytic domain reveals a novel serine protease fold and catalytic triad.Cell 86 (1996) 477-483. [PMID: 8797829]
2. Darke, P.L. Herpesvirus assemblin. In: Handbook of Proteolytic Enzymes, (Barrett, A.J., Rawlings, N.D. and Woessner, J.F. eds), pp. 470-472 (1998) Academic Press, London
[EC 3.4.21.97 created 2000]
EC 3.4.21.98
Accepted name: hepacivirin
Reaction: Hydrolysis of four peptide bonds in the viral precursor polyprotein, commonly with Asp or Glu in the P6 position, Cys or Thr in P1 and Ser or Ala in P1'
Other names: Cpro-2; hepatitis C virus NS3 serine proteinase; NS3-4A serine proteinase complex
Comments: Encoded by the genome of the viruses of the hepatitis C group, and contributes to the maturation of the precursor polyproteins. The enzyme is greatly activated by binding of the 54-residue NS4A 'cofactor' protein also derived from the viral polyprotein. Type example of peptidase family S29. The crystallographic structure shows a chymotrypsin-like fold
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 149885-80-3
References
1. Kim, J.L., Morgenstern, K.A., Lin, C., Fox, T., Dwyer, M.D., Landro, J.A., Chambers, S.P., Markland, W., Lepre, C.A., O'Malley, E.T., Harbeson, S.L., Rice, C.M., Murcko, M.A., Caron, P.R. and Thomson, J.A. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87 (1996) 343-355. [PMID: 8861917]
2. Rice, C.M. Hepatitis C virus polyprotein peptidase. In: : Handbook of Proteolytic Enzymes (Barrett, A.J., Rawlings, N.D. and Woessner, J.F. eds), pp. 272-277 (1998) Academic Press, London
[EC 3.4.21.98 created 2000]
EC 3.4.21.99
Accepted name: spermosin
Reaction: Hydrolyses arginyl bonds, preferably with Pro in the P2 position
Comments: The enzyme from the ascidian (Prochordate) Halocynthia roretzi is localised in the sperm head, and released during sperm activation. A proline-rich region is involved in binding to the vitelline coat of the egg. Belongs In peptidase family S1 (trypsin family).
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 89925-67-7
References:
1. Sawada, H., Yokosawa, H. and Ishii, S. Purification and characterization of two types of trypsin-like enzymes from sperm of the ascidian (Prochordata) Halocynthia roretzi. Evidence for the presence of spermosin, a novel acrosin-like enzyme. J. Biol. Chem. 259 (1984) 2900-2904. [PMID: 6365918]
2. Sawada, H., Yokosawa, H., Someno, T., Saino, T. and Ishii, S. Evidence for the participation of two sperm proteases, spermosin and acrosin, in fertilization of the ascidian, Halocynthia roretzi: inhibitory effects of leupeptin analogs on enzyme activities and fertilization. Dev. Biol. 105 (1984) 246-249. [PMID: 6381175]
3. Sawada, H., Iwasaki, K., Kihara-Negishi, F., Ariga, H. and Yokosawa, H. Localization, expression, and the role in fertilization of spermosin, an ascidian sperm trypsin-like protease. Biochem. Biophys. Res. Commun. 222 (1996) 499-504. [PMID: 8670234]
4. Sawada, H. and Someno, T. Substrate specificity of ascidian sperm trypsin-like proteases, spermosin and acrosin. Mol. Reprod. Dev. 45 (1996) 240-243. [PMID: 8914083]
[EC 3.4.21.99 created 2001]
EC 3.4.21.100
Accepted name: sedolisin
Reaction: Hydrolysis of the B chain of insulin at -Glu13 Ala-, -Leu15
Ala-, -Leu15 Tyr- and -Phe25
Tyr- and -Phe25 Tyr-, and angiotensin I at -Tyr4
Tyr-, and angiotensin I at -Tyr4 Ile-. A good synthetic substrate is Lys-Pro-Ile-Glu-Phe
Ile-. A good synthetic substrate is Lys-Pro-Ile-Glu-Phe Phe(NO2)-Arg-Leu.
Phe(NO2)-Arg-Leu.
Other name(s): Pseudomonas sp. pepstatin-insensitive carboxyl proteinase; pseudomonapepsin; pseudomonalisin; sedolysin
Comments: An enzyme secreted by Pseudomonas sp. No. 101. Optimum pH is 4. It is distinguished from xanthomonapepsin by its insensitivity to EPNP and from scytalidopepsin B by this property and by its unrelated amino-acid sequence. Inhibited by tyrostatin, a peptide aldehyde [2]. Type example of peptidase family S53.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 848318-58-1
References:
1. Oda, K., Sugitani, M., Fukuhara, K. and Murao, S. Purification and properties of a pepstatin-insensitive carboxyl proteinase from a Gram-negative bacterium. Biochim. Biophys. Acta 923 (1987) 463-469. [PMID: 3548827]
2. Oda, K., Nakatani, H. and Dunn, B.M. Substrate specificity and kinetic properties of pepstatin-insensitive carboxyl proteinase from Pseudomonas sp. No. 101. Biochim. Biophys. Acta 1120 (1992) 208-214. [PMID: 1562589]
3. Wlodawer, A., Li, M., Dauter, Z., Gustchina, A., Uchida, K., Oyama, H., Dunn, B.M. and Oda, K. Carboxyl proteinase from Pseudomonas defines a novel family of subtilisin-like enzymes. Nat. Struct. Biol. 8 (2001) 442-446. [PMID: 11323721]
4. Wlodawer, A., Li, M., Gustchina, A., Oyama, H., Dunn, B.M. and Oda, K. Structural and enzymatic properties of the sedolisin family of serine-carboxyl peptidases. Acta Biochim. Pol. 50 (2003) 81-102. [PMID: 12673349]
[EC 3.4.21.100 created 1995 as EC 3.4.23.37, transferred 2001 to EC 3.4.21.100, modified 2003]
Continued with EC 3.4.21.101-123
Return to Peptidases Home Page.
Return to Enzymes Home Page.